Publications
You can find the complete list of our published work in the IOCB publication database. For the full list including pre-prints, refer to the Google Scholar page.
In our lab, we conduct interdisciplinary research that combines biochemistry, metabolomics, machine learning, and synthetic biology. Below, we selected a few representative publications from our research in each of the aforementioned fields.
Exploring biosynthesis & chemodiversity of natural products
- We study biosyntheses in selected plant families and the chemodiversity of their natural products.
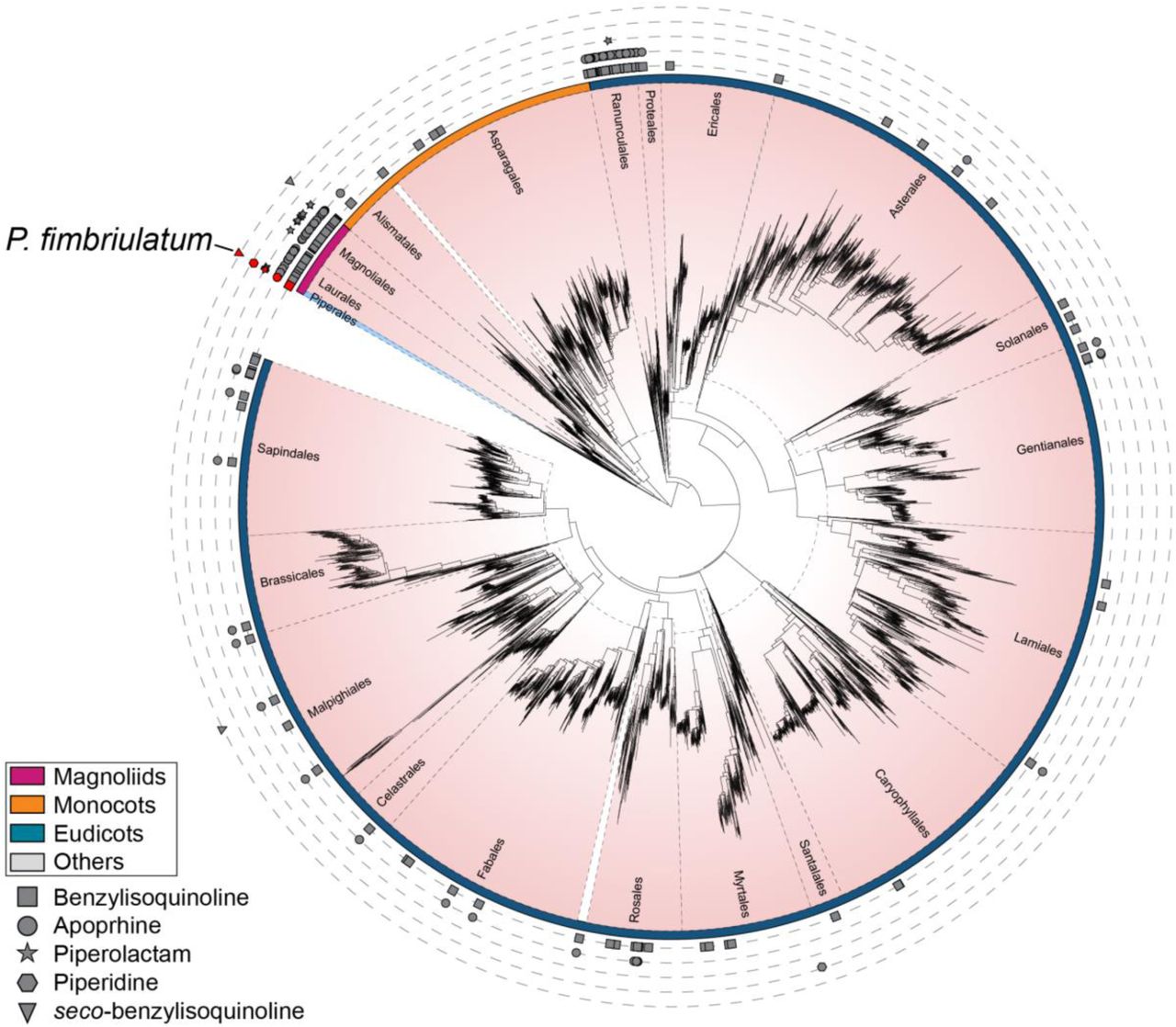
Computational metabolomics reveals overlooked chemodiversity of alkaloid scaffolds in Piper fimbriulatum.
Damiani, T., Smith, J., Hebra, T., Perkovic, M., Cicak, M., Kadlecova, A., ... & Pluskal, T.
The Plant Journal, 2025.
Studying Plant Specialized Metabolites Using Computational Metabolomics Strategies.
Mutabdžija, L., Myoli, A., de Jonge, N. F., Damiani, T., Schmid, R., van der Hooft, J. J., ... & Pluskal, T.
Plant Functional Genomics: Methods and Protocols, 2024.
The biosynthetic origin of psychoactive kavalactones in kava.
Pluskal, T., Torrens-Spence, M. P., Fallon, T. R., De Abreu, A., Shi, C. H., & Weng, J. K.
Nature Plants, 2019.
Computational metabolomics
- We develop advanced computational tools to enhance the identification and interpretation of mass spectrometry (MS) data.
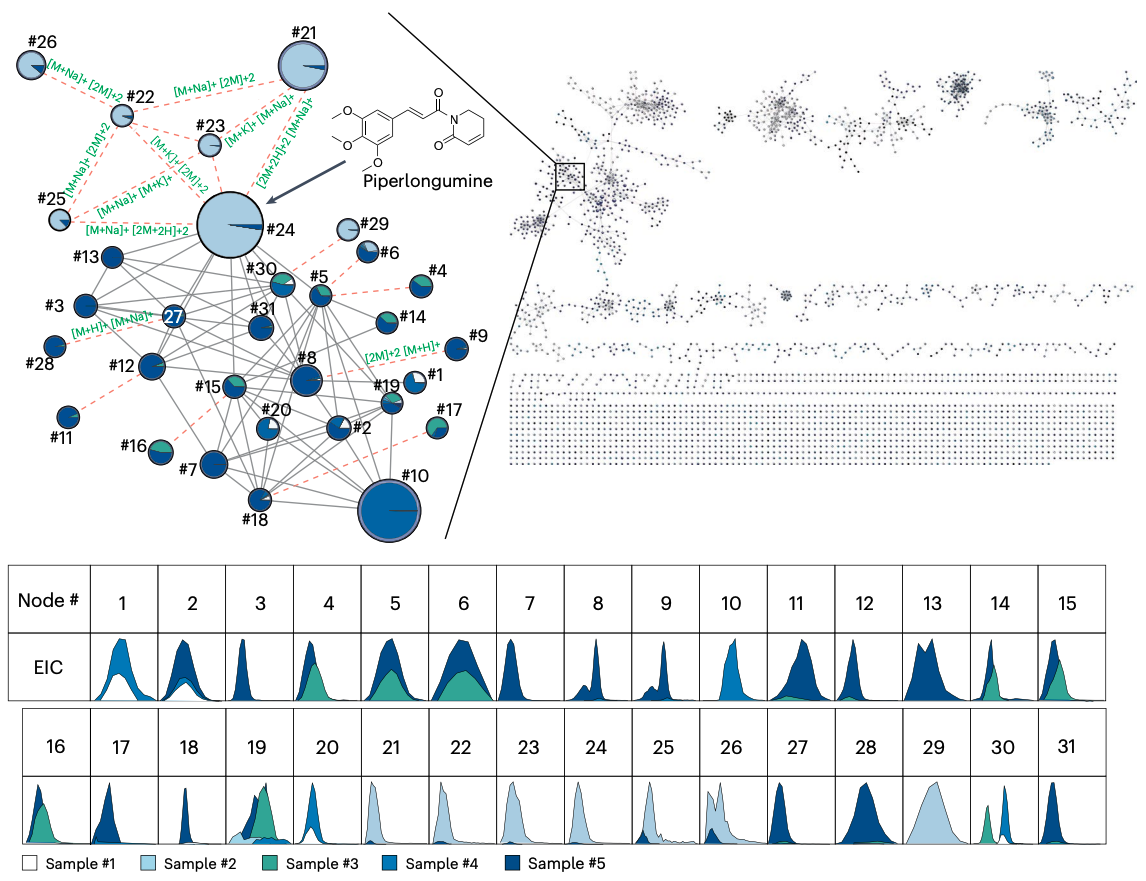
Statistical analysis of feature-based molecular networking results from non-targeted metabolomics data.
Pakkir Shah, A. K., Walter, A., Ottosson, F., Russo, F., Navarro-Diaz, M., Boldt, J., ... & Petras, D.
Nature Protocols, 2025.
MassSpecGym: A benchmark for the discovery and identification of molecules.
Bushuiev, R., Bushuiev, A., de Jonge, N., Young, A., Kretschmer, F., Samusevich, R., ... & Pluskal, T.
Advances in Neural Information Processing Systems, 2025.
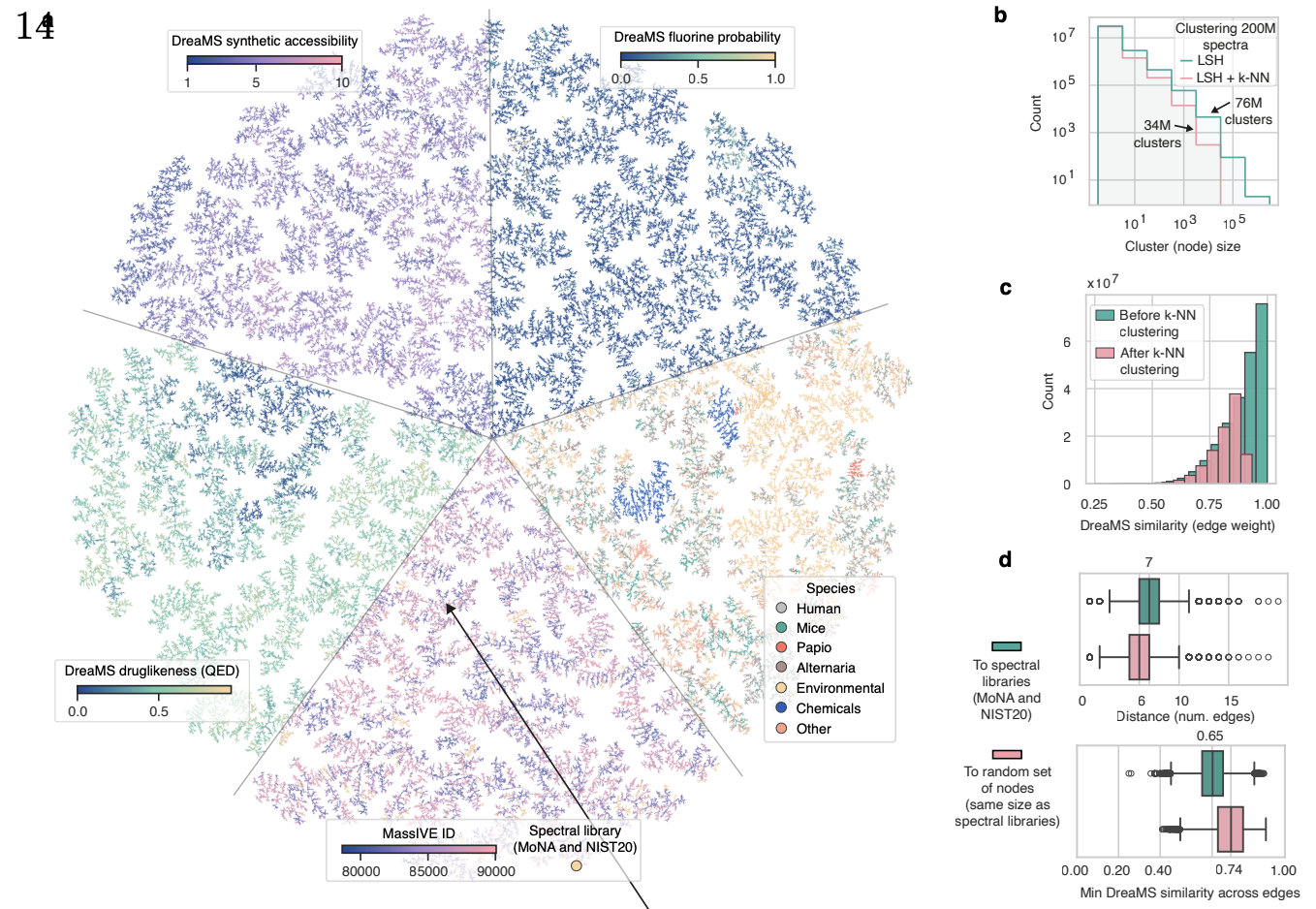
Emergence of molecular structures from repository-scale self-supervised learning on tandem mass spectra.
Bushuiev, R., Bushuiev, A., Samusevich, R., Brungs, C., Sivic, J., & Pluskal, T.
Nature Biotechnology (in press), 2025.
Efficient generation of open multi-stage fragmentation mass spectral libraries.
Brungs, C., Schmid, R., Heuckeroth, S., Mazumdar, A., Drexler, M., Šácha, P., ... & Pluskal, T.
Nature Methods (in press), 2024.
Reproducible mass spectrometry data processing and compound annotation in MZmine 3.
Heuckeroth, S., Damiani, T., Smirnov, A., Mokshyna, O., Brungs, C., Korf, A., ... & Pluskal, T.
Integrative analysis of multimodal mass spectrometry data in MZmine 3.
Schmid, R., Heuckeroth, S., Korf, A., Smirnov, A., Myers, O., Dyrlund, T. S., ... & Pluskal, T.
Nature Biotechnology, 2023.
Machine learning powered enzyme development
- We are working towards controlled engineering of novel enzymes with desired enzymatic activity by leveraging state-of-the-art machine learning methods.
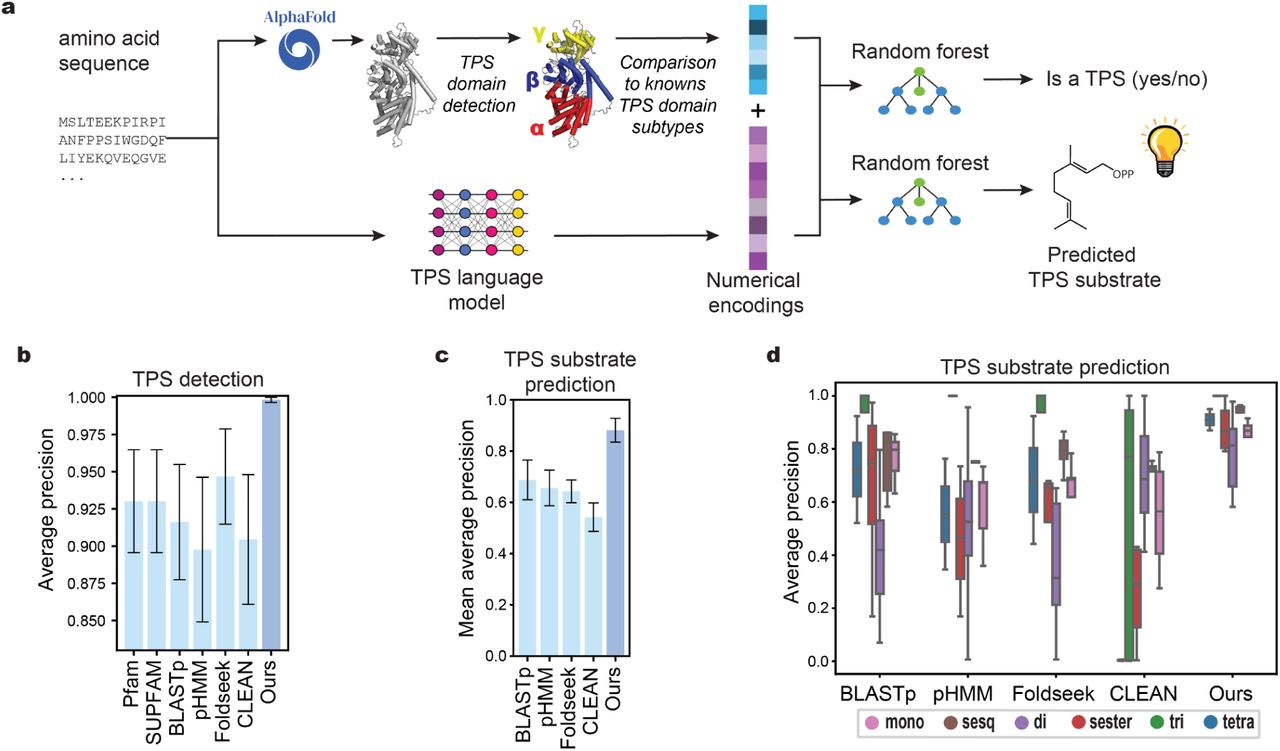
Highly accurate discovery of terpene synthases powered by machine learning reveals functional terpene cyclization in Archaea.
Samusevich, R., Hebra, T., Bushuiev, R., Bushuiev, A., Čalounová, T., Smrčková, H., ... & Pluskal, T.
bioRxiv, 2024.
Revealing data leakage in protein interaction benchmarks.
Bushuiev, A., Bushuiev, R., Sedlar, J., Pluskal, T., Damborsky, J., Mazurenko, S., & Sivic, J.
International Conference on Learning Representations, 2024.
Learning to design protein-protein interactions with enhanced generalization.
Bushuiev, A., Bushuiev, R., Kouba, P., Filkin, A., Gabrielova, M., Gabriel, M., ... & Sivic, J.
International Conference on Learning Representations, 2024.
Machine learning-guided protein engineering.
Kouba, P., Kohout, P., Haddadi, F., Bushuiev, A., Samusevich, R., Sedlar, J., ... & Mazurenko, S.
ACS catalysis, 2023.
Synthetic biology
- We use synthetic biology methods and develop novel tools to advance the field.
POMBOX: A Fission Yeast Cloning Toolkit for Molecular and Synthetic Biology.
T Hebra, H Smrčková, B Elkatmis, M Prevorovsky, T Pluskal.
ACS Synthetic Biology, 2023. Cover Article.