Our projects
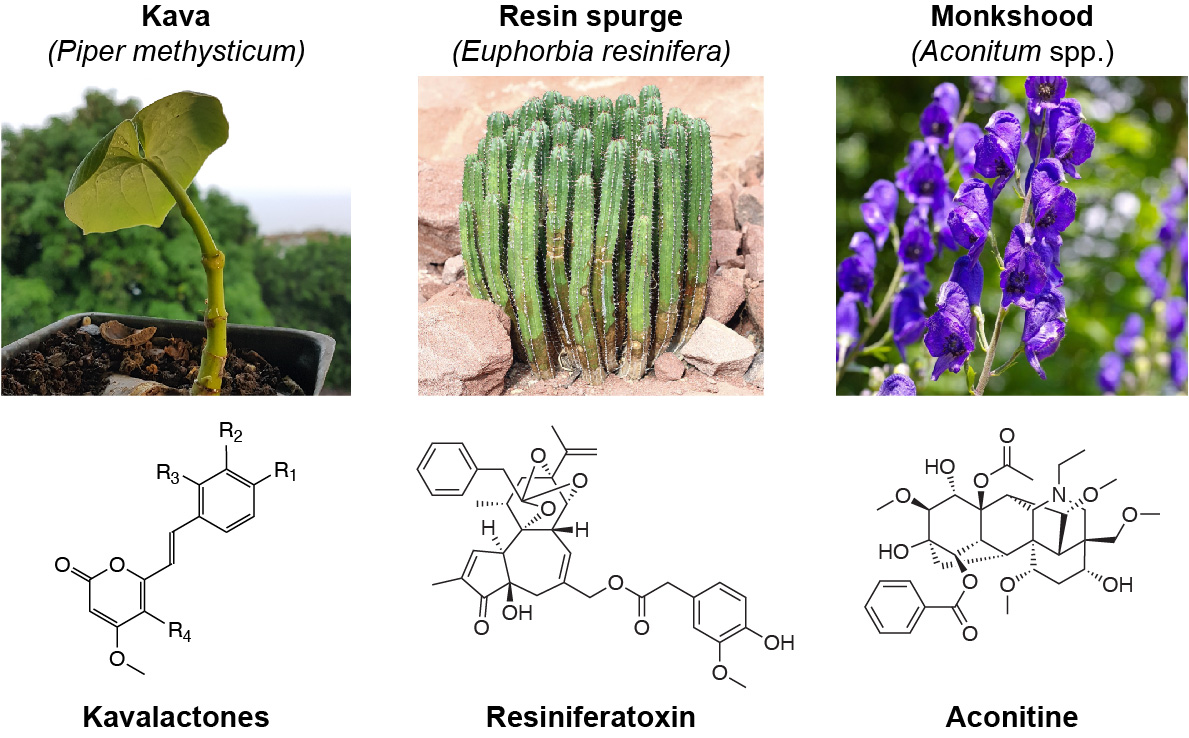
During millions of years of evolution, land plants have created an astonishing variety of bioactive specialized metabolites (also referred to as ‘secondary metabolites’ or ‘natural products’) to support their defense and ecological adaptation. Such molecules often interact with human molecular receptors, thus providing an essential source of chemical scaffolds for the development of new medicines. About 25% of prescription drugs currently in use originated from plants.
The structural and stereochemical complexity of plant metabolites often renders their chemical synthesis unfeasible, but recent progress in sequencing and metabolomics technologies has opened a new avenue towards heterologous production of plant metabolites using their native biosynthetic enzymes. However, the discovery and engineering of metabolic pathways from plants remains very difficult. Our lab combinines novel computational (e.g., machine learning) and experimental approaches to develop rapid, generally applicable workflows for the discovery and utilization of bioactive molecules derived from plants.
Exploring biosynthesis & chemodiversity of natural products
Computational metabolomics
In our lab, we have developed advanced computational tools to enhance the identification and interpretation of mass spectrometry (MS) data. We focus on machine-learning approaches and scalable data analysis platforms to improve metabolite annotation and molecular structure elucidation.
Machine learning powered enzyme development
Currently, it is impossible to accurately predict the functions of newly discovered enzymes directly from their amino acid sequence or to generate enzymes for specific reactions. We are capable of creating new versions of known enzymes, but no one has yet managed to develop entirely new enzymes for reactions we’ve never seen before.
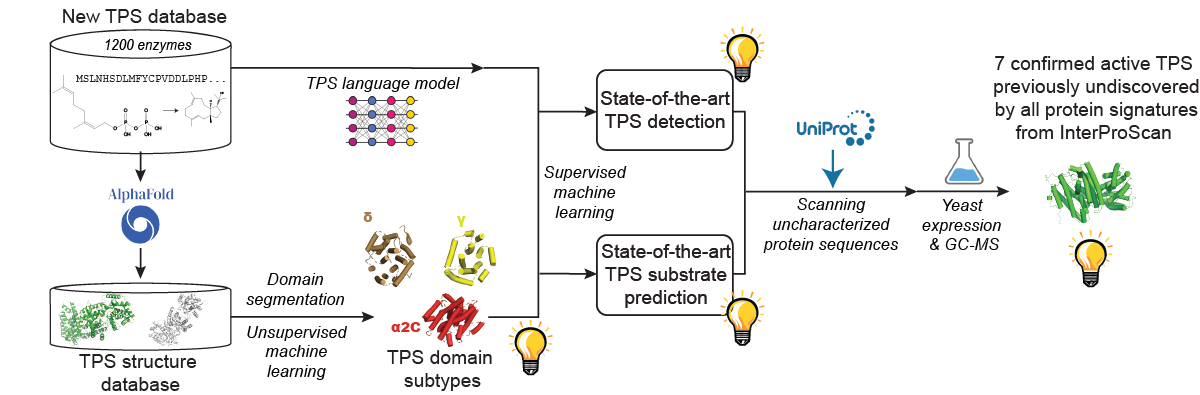
We are working to overcome this biotechnology bottleneck by developing machine learning models for the prediction of functions and de novo generation of a single well-defined class of enzymes - terpene synthases (TPSs). TPSs are ubiquitous enzymes that produce the core hydrocarbon scaffolds for the largest and the most diverse class of natural products called terpenoids. Our lab curates a freely available database of terpene synthases and their reaction mechanisms.
To further this goal, we’ve developed Enzyme Explorer, a state-of-the-art machine-learning pipeline for automated TPS detection and substrate prediction. From the protein sequence only, our method can detect TPSs and their substrates with high accuracy, even for rare classes of TPSs. By leveraging our predictive pipeline, we were the first to report three experimentally confirmed active TPSs in Archaea.
Bioinformatics projects
Our lab maintains the following bioinformatics projects:
We regularly participate in the Google Summer of Code program!
Other projects
Students from our lab founded the first Prague-based team to take part in the iGEM 2025 competition with their own innovative project!




